- Overview
- Installation
- Configuration
- Example models
- API reference
- Tips and tricks
- Hardware setup
Integrator¶
This demo implements a one-dimensional neural integrator.
This example utilizes a recurrent network. It shows how neurons can be used to implement stable dynamics. Such dynamics are important for memory, noise cleanup, statistical inference, and many other dynamic transformations.
[1]:
import matplotlib.pyplot as plt
%matplotlib inline
import nengo
from nengo.processes import Piecewise
import nengo_loihi
nengo_loihi.set_defaults()
/home/travis/build/nengo/nengo-loihi/nengo_loihi/version.py:23: UserWarning: This version of `nengo_loihi` has not been tested with your `nengo` version (3.0.1.dev0). The latest fully supported version is 3.0.0
"supported version is %s" % (nengo.__version__, latest_nengo_version)
/home/travis/virtualenv/python3.6.3/lib/python3.6/site-packages/nengo_dl/version.py:42: UserWarning: This version of `nengo_dl` has not been tested with your `nengo` version (3.0.1.dev0). The latest fully supported version is 3.0.0.
% ((nengo.version.version,) + latest_nengo_version)
Creating the network in Nengo¶
Our model consists of one recurrently connected ensemble, and an input node. The input node will provide a piecewise step function as input so that we can see the effects of recurrence.
[2]:
with nengo.Network(label='Integrator') as model:
ens = nengo.Ensemble(n_neurons=120, dimensions=1)
stim = nengo.Node(
Piecewise({
0: 0,
0.2: 1,
1: 0,
2: -2,
3: 0,
4: 1,
5: 0
}))
# Connect the population to itself
tau = 0.1
nengo.Connection(ens, ens,
transform=[[1]],
synapse=tau)
nengo.Connection(
stim, ens, transform=[[tau]], synapse=tau)
# Collect data for plotting
stim_probe = nengo.Probe(stim)
ens_probe = nengo.Probe(ens, synapse=0.01)
Running the network in Nengo¶
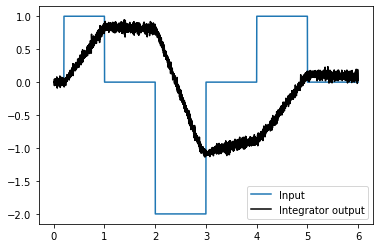
We can use Nengo to see the desired model output.
[3]:
with nengo.Simulator(model) as sim:
sim.run(6)
t = sim.trange()
0%
0%
[4]:
def plot_decoded(t, data):
plt.figure()
plt.plot(t, data[stim_probe], label="Input")
plt.plot(t, data[ens_probe], 'k', label="Integrator output")
plt.legend()
plot_decoded(t, sim.data)
Running the network with Nengo Loihi¶
[5]:
with nengo_loihi.Simulator(model) as sim:
sim.run(6)
t = sim.trange()
/home/travis/build/nengo/nengo-loihi/nengo_loihi/discretize.py:471: UserWarning: Lost 2 extra bits in weight rounding
warnings.warn("Lost %d extra bits in weight rounding" % (-s2,))
[6]:
plot_decoded(t, sim.data)
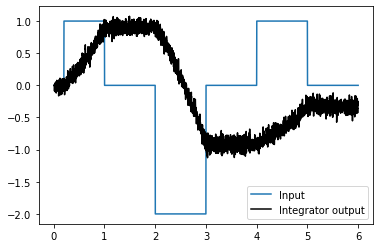
The network integrates its input, but without input decays quicker than the Nengo model. Likely the same workarounds discussed in the communication channel example will be useful here.