Controlled question answering¶
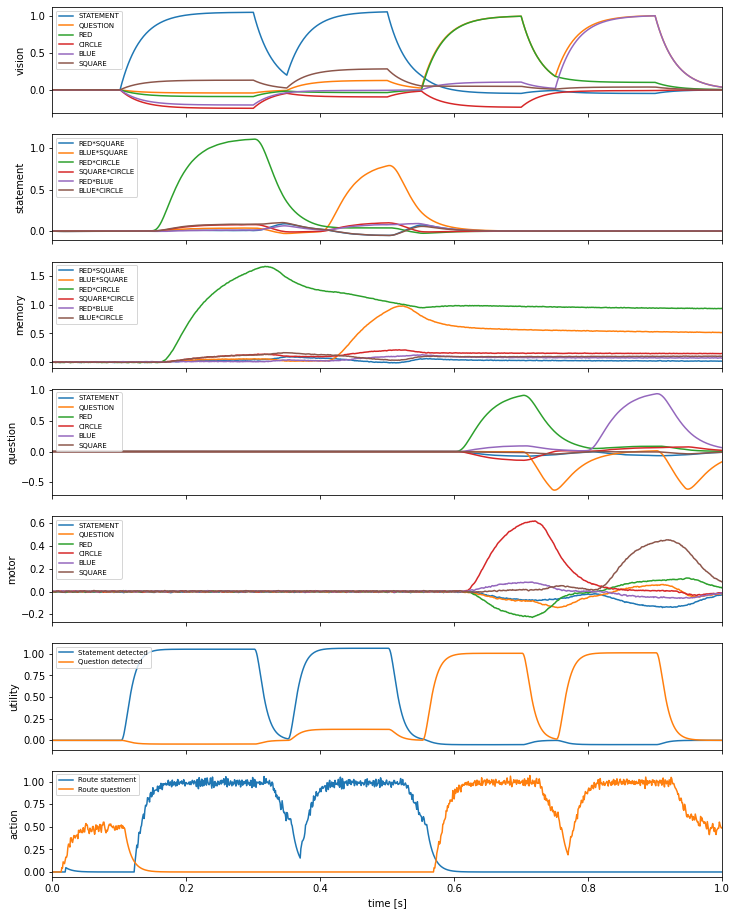
This demo implements a simple form of question answering by using the basal ganglia to store and retrieve information from working memory in response to visual input. More specifically, the basal ganglia decides what to do with the information in the visual channel based on its content (i.e. whether it is a statement or a question)
When you run the network, it will start by binding RED and CIRCLE and then binding BLUE and SQUARE so the memory essentially has RED * CIRCLE + BLUE * SQUARE. It does this because it is told that RED * CIRCLE is a STATEMENT (i.e. RED * CIRCLE + STATEMENT in the code) as is BLUE * SQUARE. Then it is presented with something like QUESTION + RED (i.e., “What is red?”). The basal ganglia then reroutes that input to be compared to what is in working memory and the
result shows up in the motor channel.
[1]:
%matplotlib inline
import matplotlib.pyplot as plt
import nengo
import nengo_spa as spa
seed = 0
Create the model¶
Notice that when you specify actions, you’re determining which modules are connected to which. For example, by having a mapping that depends on the state of cortex, you are determining that the cortex and basal ganglia must be connected. As well, when you specify that the result of the action changes the state of cortex, then you are determining that thalamus must be connected to cortex.
[2]:
def input_fn(t):
if 0.1 < t < 0.3:
return "STATEMENT+RED*CIRCLE"
elif 0.35 < t < 0.5:
return "STATEMENT+BLUE*SQUARE"
elif 0.55 < t < 0.7:
return "QUESTION+RED"
elif 0.75 < t < 0.9:
return "QUESTION+BLUE"
else:
return "0"
# Number of dimensions for the Semantic Pointers
dimensions = 128
# Number of neurons to represent each dimension
# Higher number for more accurate representation
n_per_dim = 100
# Make a model object with the SPA network
model = spa.Network(label="Controlled Question Answering", seed=seed)
with model:
# Specify the modules to be used
vision = spa.Transcode(input_fn, output_vocab=dimensions)
motor = spa.State(dimensions, neurons_per_dimension=n_per_dim)
memory = spa.State(
dimensions, neurons_per_dimension=n_per_dim, feedback=1.0, feedback_synapse=0.1
)
# Transcode modules to show intermediate results
statement = spa.Transcode(input_vocab=dimensions, output_vocab=dimensions)
question = spa.Transcode(input_vocab=dimensions, output_vocab=dimensions)
# Specify the action mapping
with spa.ActionSelection() as action_sel:
spa.ifmax(
spa.dot(vision, spa.sym.STATEMENT),
vision - spa.sym.STATEMENT >> statement,
statement >> memory,
)
spa.ifmax(
spa.dot(vision, spa.sym.QUESTION),
vision - spa.sym.QUESTION >> question,
~(question) * memory >> motor,
)
Probe the output¶
[3]:
with model:
p_vision = nengo.Probe(vision.output, synapse=0.03)
p_statement = nengo.Probe(statement.input, synapse=0.03)
p_question = nengo.Probe(question.input, synapse=0.03)
p_motor = nengo.Probe(motor.output, synapse=0.03)
p_memory = nengo.Probe(memory.output, synapse=0.03)
p_selected_actions = nengo.Probe(action_sel.thalamus.output, synapse=0.01)
p_utility = nengo.Probe(action_sel.bg.input, synapse=0.01)
Run the model¶
[4]:
with nengo.Simulator(model) as sim:
sim.run(1.0)
Plot the results¶
[5]:
vocab = model.vocabs[dimensions]
fig, axes = plt.subplots(7, sharex=True, figsize=(12, 16))
axes[0].plot(sim.trange(), spa.similarity(sim.data[p_vision], vocab))
axes[0].legend(vocab.keys(), fontsize="x-small", loc="upper left")
axes[0].set_ylabel("vision")
attrs = ("RED", "BLUE", "SQUARE", "CIRCLE")
attr_vecs = vocab.create_subset(attrs)
attr_pairs = spa.pairs(attr_vecs)
attr_pair_vecs = [vocab.parse(p).v for p in attr_pairs]
axes[1].plot(sim.trange(), spa.similarity(sim.data[p_statement], attr_pair_vecs))
axes[1].legend(attr_pairs, fontsize="x-small", loc="upper left")
axes[1].set_ylabel("statement")
axes[2].plot(sim.trange(), spa.similarity(sim.data[p_memory], attr_pair_vecs))
axes[2].legend(attr_pairs, fontsize="x-small", loc="upper left")
axes[2].set_ylabel("memory")
axes[3].plot(sim.trange(), spa.similarity(sim.data[p_question], vocab))
axes[3].legend(vocab.keys(), fontsize="x-small", loc="upper left")
axes[3].set_ylabel("question")
axes[4].plot(sim.trange(), spa.similarity(sim.data[p_motor], vocab))
axes[4].legend(vocab.keys(), fontsize="x-small", loc="upper left")
axes[4].set_ylabel("motor")
axes[5].plot(sim.trange(), sim.data[p_utility])
axes[5].legend(
("Statement detected", "Question detected"), fontsize="x-small", loc="upper left"
)
axes[5].set_ylabel("utility")
axes[6].plot(sim.trange(), sim.data[p_selected_actions])
axes[6].legend(
("Route statement", "Route question"), fontsize="x-small", loc="upper left"
)
axes[6].set_ylabel("action")
axes[6].set_xlabel("time [s]")
axes[6].set_xlim((0, 1.0))
[5]:
(0.0, 1.0)