- NengoLoihi
- Examples
- Communication channel
- Integrator
- Multidimensional integrator
- Simple oscillator
- Nonlinear oscillator
- Neuron to neuron connections
- PES learning
- Keyword spotting
- MNIST convolutional network
- CIFAR-10 convolutional network
- Converting a Keras model to an SNN on Loihi
- Nonlinear adaptive control
- Legendre Memory Units on Loihi
- DVS from file
- Requirements
- License
- Examples
- How to Build a Brain
- NengoCore
- Ablating neurons
- Deep learning
Neuron to neuron connections¶
While Nengo is often used with deep learning and NEF style networks, it can also be used for lower level models in which each neuron to neuron connection is explicitly specified.
In these examples, we connect a pre population to a post population with different sets of specified connection weights.
[1]:
import matplotlib.pyplot as plt
%matplotlib inline
import numpy as np
import nengo
from nengo.utils.matplotlib import rasterplot
import nengo_loihi
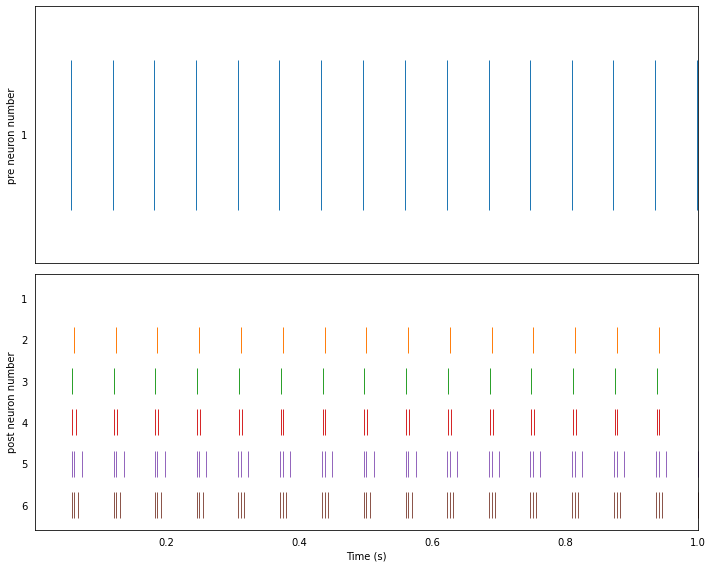
1. Simple fan-out¶
In this example, a neuron is connected to several downstream neurons with increasing synaptic strength.
Synaptic strengths are defined through the transform of a nengo.Connection. While a Connection between two ensembles operates on their vector representations, a connection between two ensemble’s neuron values operates directly on neural activities (i.e., spikes).
[2]:
with nengo.Network() as model:
pre = nengo.Ensemble(1, dimensions=1, gain=[1], bias=[1.05])
post = nengo.Ensemble(6, dimensions=1, gain=np.ones(6), bias=np.zeros(6))
transform = np.linspace(0.01, 0.15, post.n_neurons)
transform = transform.reshape((post.n_neurons, pre.n_neurons))
nengo.Connection(pre.neurons, post.neurons, transform=transform)
pre_probe = nengo.Probe(pre.neurons)
post_probe = nengo.Probe(post.neurons)
Running the network in Nengo¶
[3]:
with nengo.Simulator(model) as sim:
sim.run(1)
t = sim.trange()
/home/tbekolay/Code/nengo/nengo/neurons.py:584: RuntimeWarning: divide by zero encountered in log1p
self.tau_ref - self.tau_rc * np.log1p(1.0 / (gain * (intercepts - 1) - 1))
[4]:
def plot_rasters(t, data):
plt.figure(figsize=(10, 8))
plt.subplot(2, 1, 1)
rasterplot(t, data[pre_probe])
plt.xticks(())
plt.ylabel("pre neuron number")
plt.subplot(2, 1, 2)
rasterplot(t, data[post_probe])
plt.ylabel("post neuron number")
plt.xlabel("Time (s)")
plt.tight_layout()
plot_rasters(t, sim.data)
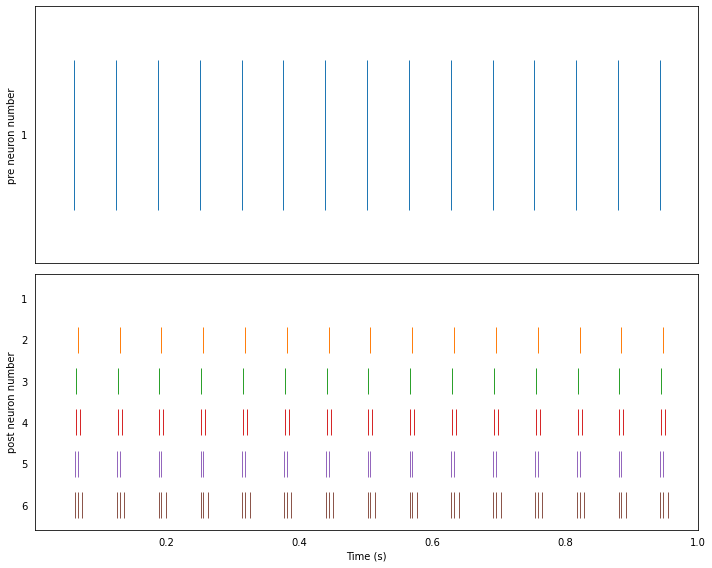
Running the network with NengoLoihi¶
[5]:
with nengo_loihi.Simulator(model) as sim:
sim.run(1)
t = sim.trange()
/home/tbekolay/Code/nengo-loihi/nengo_loihi/builder/ensemble.py:164: UserWarning: NengoLoihi does not support initial values for 'voltage' being non-zero on LIF neurons. On the chip, all values will be initialized to zero.
warnings.warn(
/home/tbekolay/Code/nengo/nengo/neurons.py:584: RuntimeWarning: divide by zero encountered in log1p
self.tau_ref - self.tau_rc * np.log1p(1.0 / (gain * (intercepts - 1) - 1))
[6]:
plot_rasters(t, sim.data)
2. One-to-one connections¶
In this example, two populations of equal size are connected one-to-one with random biases in the pre population and random excitatory connection weights.
[7]:
rng = np.random.RandomState(seed=10)
n_neurons = 5
with nengo.Network() as model:
pre = nengo.Ensemble(
n_neurons,
1,
gain=np.ones(n_neurons),
bias=rng.uniform(low=1.0, high=1.5, size=n_neurons),
)
post = nengo.Ensemble(
n_neurons, 1, gain=np.ones(n_neurons), bias=np.zeros(n_neurons)
)
transform = np.zeros((n_neurons, n_neurons))
di = np.diag_indices(n_neurons)
transform[di] = rng.uniform(low=0.0, high=0.2, size=n_neurons)
nengo.Connection(pre.neurons, post.neurons, transform=transform)
pre_probe = nengo.Probe(pre.neurons)
post_probe = nengo.Probe(post.neurons)
Running the network in Nengo¶
[8]:
with nengo.Simulator(model) as sim:
sim.run(1)
t = sim.trange()
plot_rasters(t, sim.data)
/home/tbekolay/Code/nengo/nengo/neurons.py:584: RuntimeWarning: divide by zero encountered in log1p
self.tau_ref - self.tau_rc * np.log1p(1.0 / (gain * (intercepts - 1) - 1))
Running the network with NengoLoihi¶
[9]:
with nengo_loihi.Simulator(model) as sim:
sim.run(1)
t = sim.trange()
plot_rasters(t, sim.data)
/home/tbekolay/Code/nengo-loihi/nengo_loihi/builder/ensemble.py:164: UserWarning: NengoLoihi does not support initial values for 'voltage' being non-zero on LIF neurons. On the chip, all values will be initialized to zero.
warnings.warn(
/home/tbekolay/Code/nengo/nengo/neurons.py:584: RuntimeWarning: divide by zero encountered in log1p
self.tau_ref - self.tau_rc * np.log1p(1.0 / (gain * (intercepts - 1) - 1))
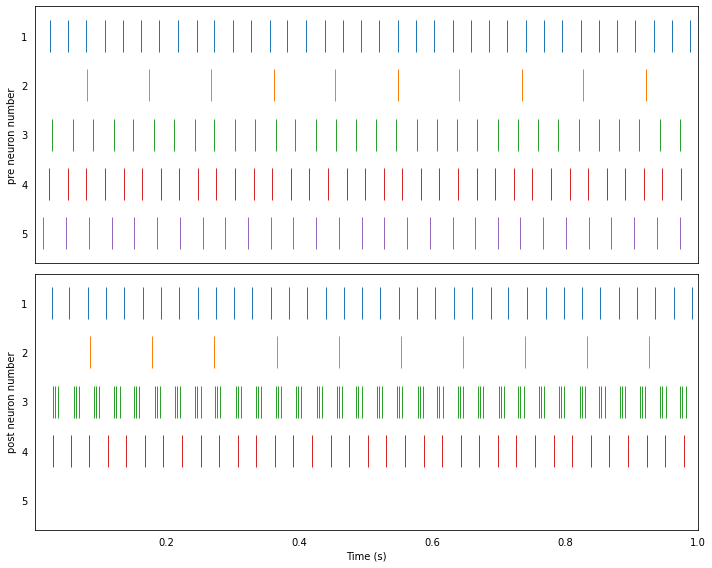
3. Fixed probability connections¶
In this example, two populations are recurrently connected (i.e., post is also connected back to pre). There is a fixed probability of two neurons being connected in either direction, a fixed probability of an inhibitory connection, and all connections have the same weight.
[10]:
rng = np.random.RandomState(seed=100)
inhibitory = 0.5 # 50% inhibitory connections
connection_prob = 0.6 # 60% probability of being connected
n_neurons = 25
with nengo.Network() as model:
pre = nengo.Ensemble(
n_neurons,
1,
gain=np.ones(n_neurons),
bias=rng.uniform(low=-2, high=2, size=n_neurons),
)
post = nengo.Ensemble(
n_neurons,
1,
gain=np.ones(n_neurons),
bias=rng.uniform(low=-2, high=2, size=n_neurons),
)
pre_post = np.ones((n_neurons, n_neurons)) * 0.05
# Make some inhibitory
pre_post[rng.rand(n_neurons, n_neurons) <= inhibitory] *= -1
# Remove 1 - connection_prob connections
pre_post[rng.rand(n_neurons, n_neurons) > connection_prob] = 0
nengo.Connection(pre.neurons, post.neurons, transform=pre_post)
post_pre = np.ones((n_neurons, n_neurons)) * 0.05
post_pre[rng.rand(n_neurons, n_neurons) <= inhibitory] *= -1
post_pre[rng.rand(n_neurons, n_neurons) > connection_prob] = 0
nengo.Connection(post.neurons, pre.neurons, transform=post_pre)
pre_probe = nengo.Probe(pre.neurons)
post_probe = nengo.Probe(post.neurons)
Running the network in Nengo¶
[11]:
with nengo.Simulator(model) as sim:
sim.run(1)
t = sim.trange()
plot_rasters(t, sim.data)
/home/tbekolay/Code/nengo/nengo/neurons.py:584: RuntimeWarning: invalid value encountered in log1p
self.tau_ref - self.tau_rc * np.log1p(1.0 / (gain * (intercepts - 1) - 1))
/home/tbekolay/Code/nengo/nengo/neurons.py:587: UserWarning: Non-finite values detected in `max_rates`; this probably means that `gain` was too small.
warnings.warn(

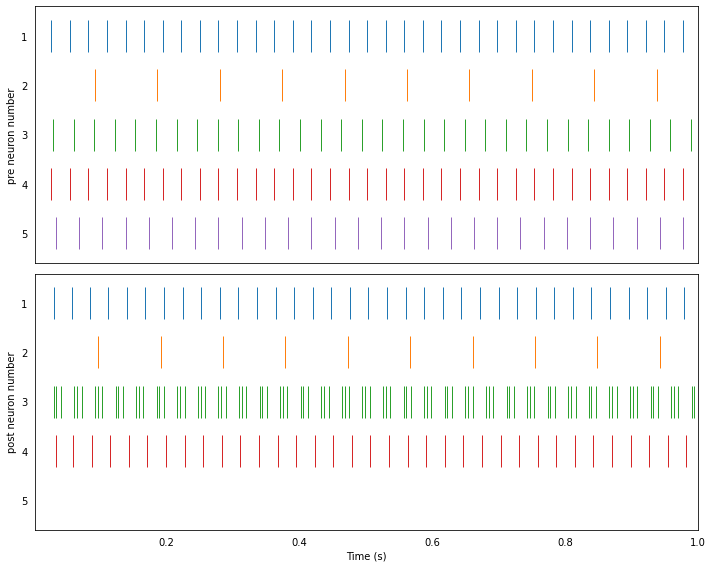
Running the network with NengoLoihi¶
[12]:
with nengo_loihi.Simulator(model) as sim:
sim.run(1)
t = sim.trange()
plot_rasters(t, sim.data)
/home/tbekolay/Code/nengo/nengo/neurons.py:584: RuntimeWarning: invalid value encountered in log1p
self.tau_ref - self.tau_rc * np.log1p(1.0 / (gain * (intercepts - 1) - 1))
/home/tbekolay/Code/nengo/nengo/neurons.py:587: UserWarning: Non-finite values detected in `max_rates`; this probably means that `gain` was too small.
warnings.warn(
/home/tbekolay/Code/nengo-loihi/nengo_loihi/builder/ensemble.py:164: UserWarning: NengoLoihi does not support initial values for 'voltage' being non-zero on LIF neurons. On the chip, all values will be initialized to zero.
warnings.warn(
