- NengoLoihi
- How to Build a Brain
- Tutorials
- A Single Neuron Model
- Representing a scalar
- Representing a Vector
- Addition
- Arbitrary Linear Transformation
- Nonlinear Transformations
- Structured Representations
- Question Answering
- Question Answering with Control
- Question Answering with Memory
- Learning a communication channel
- Sequencing
- Routed Sequencing
- Routed Sequencing with Cleanup Memory
- Routed Sequencing with Cleanup all Memory
- 2D Decision Integrator
- Requirements
- License
- Tutorials
- NengoCore
- Ablating neurons
- Deep learning
Sequencing¶
This model uses the basal ganglia model to cycle through a sequence of five representations (i.e., A -> B -> C -> D -> E -> A -> …). The model incorporates a working memory component (memory), which allows the Basal Ganglia to update that memory based on a set of condition/action mappings.
[1]:
# Setup the environment
from nengo import spa # import spa related packages
Create the model¶
The model has parameters as described in the book. In the book (Nengo 1.4) separate “Rules” and “Sequence” classes were created. However, this is not needed in Nengo 2.0 since you can directly specify the rules using the built-in “Actions” class in the nengo.spa package. This class takes a string definition of the action as an input as shown in the code where --> is used to split the action into condition and effect. If no --> is used, it is treated as having no condition and just
effect.
The syntax for creating an input function in Nengo 2.0 is also different from that in Nengo 1.4 mentioned in the book. The syntax for Nengo 2.0, which you will use here, is spa.Input(<module>=<function>). The first parameter <module> refers to name of the module that you want to provide input to and the second parameter <function> refers to the function to execute to generate inputs to that module. The functions should always return strings, which will then be parsed by the relevant
SPA vocabulary.
In Nengo 1.4, a memory element for representing the state was created by using the Buffer() object as described in the book. However, in Nengo 2.0, you will use the State() object with the feedback parameter set to 1 for creating a memory module capable of storing a vector over time.
[2]:
# Number of dimensions for the Semantic Pointers
dim = 16
# Create the spa.SPA network to which we can add SPA objects
model = spa.SPA(label="Sequence")
with model:
# Creating a working memory/cortical element
model.state = spa.State(dimensions=dim, feedback=1, feedback_synapse=0.01)
# Specifying the action mappings (rules) for BG and Thal
actions = spa.Actions(
"dot(state, A) --> state = B",
"dot(state, B) --> state = C",
"dot(state, C) --> state = D",
"dot(state, D) --> state = E",
"dot(state, E) --> state = A",
)
# Creating the BG and thalamus components that confirm to the specified rules
model.bg = spa.BasalGanglia(actions=actions)
model.thal = spa.Thalamus(model.bg)
# Function that provides the model with an initial input semantic pointer.
def start(t):
if t < 0.1: # Duration of the initial input = 0.1
return "D"
return "0"
# Input
model.input = spa.Input(state=start)
Run the model¶
[ ]:
# Import the nengo_gui visualizer to run and visualize the model.
from nengo_gui.ipython import IPythonViz
IPythonViz(model, "ch7-spa-sequence.py.cfg")
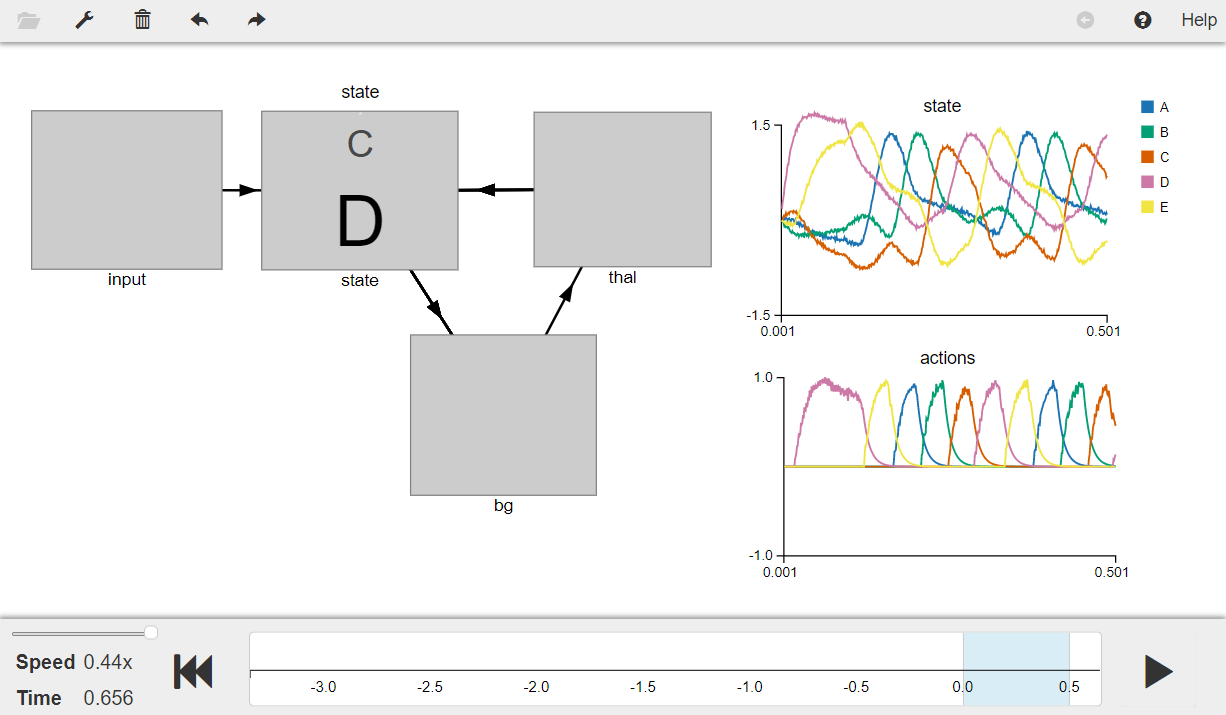
Press the play button in the visualizer to run the simulation. You should see the graphs as shown in the figure below.
The graph on the top-left shows the semantic pointer representation of the values stored in the state ensemble. The plot on the bottom-right shows the current transition or the action being executed, and the plot on the top-right shows the utility (similarity) of the current Basal Ganglia input (i.e., state) with the possible vocabulary vectors.
The book describes that the results of the model can be seen through the visualizer in Nengo 1.4 GUI which has a “Utility” box and the “Rules” box. Note that the bottom-right plot shows the same information as seen in the “Rules” box and top-right plot shows the same information as seen in the “Utility” box.
[3]:
from IPython.display import Image
Image(filename="ch7-spa-sequence.png")
[3]: